Research Projects | Overview
Defining the Human Articular Cartilage Lineage
Our lab has developed methods that enable us to reproducibly and efficiently generate two distinct cartilage lineages from human embryonic stem cells and induced pluripotent stem cells (collectively hPSCs). One chondrocyte lineage has the ability to generate stable cartilage in vivo (a function of articular cartilage) and the other produces a cartilaginous tissue that undergoes ossification (a function of growth plate cartilage to form new bone). Using a combination of functional challenge assays and bulk/single cell RNA sequencing, we are focused on understanding precisely when lineage commitment occurs and the molecular mechanism associated with these branching points/differentiation trajectories.
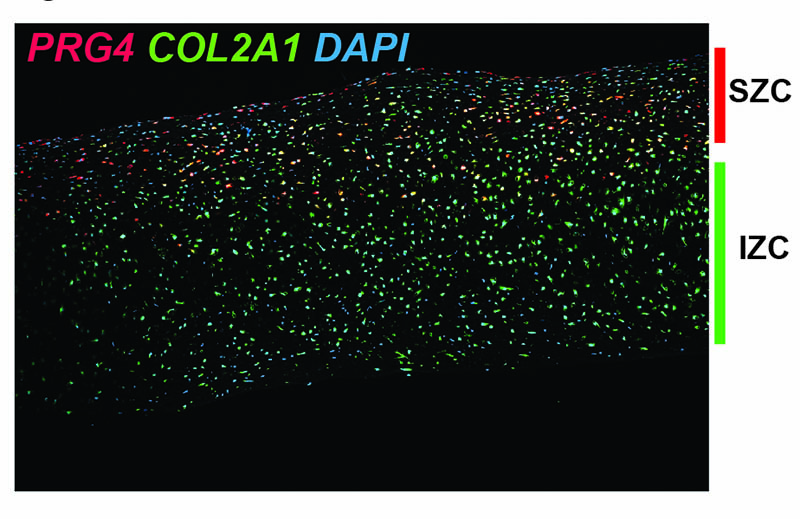
Zonal organization of hESC-derived articular cartilage
Similar to cartilage found in our joints, the flattened layer of chondrocytes within the superficial layers of the hESC-derived articular cartilage express PRG4, encoding the protein lubricin, while deeper intermediate-zone-like chondrocytes express only COL2A1, encoding type II collagen. mRNA expression detected by in situ hybridization (RNAScope).
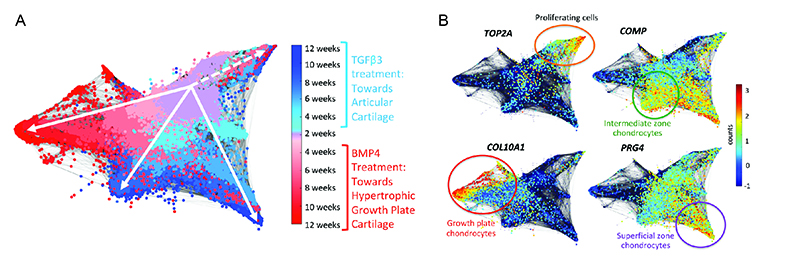
Single cell transcriptomic analyses of human chondrogenesis
Molecular analysis of hESC-derived chondrogenesis towards the articular or growth plate lineages at single cell resolution. (A) Representative STITCH plot illustrates cellular trajectories of hESC-derived chondrocytes as they differentiate into articular (blues) or growth plate-like (reds) cartilage. (B) Feature plots illustrate mRNA counts of marker genes representing distinct cartilage zones. The most mature chondrocyte cells are present at the latest timepoints (12 weeks) localized at the ends of each respective branch.
Knee in a Dish
Our approach to differentiating pluripotent stem cells is based on embryonic development, and thus it allows us to create and have access to unlimited amounts of various developmental progenitors along the way. We are interested in expanding our model of articular cartilage development to study the development of other joint tissues, as well as the early events that are associated with joint initiation in the limb bud. We have created several mouse and human ESC reporter lines to mark early joint progenitor cells (Gdf5/GDF5+) and downstream joint tissues such as articular cartilage (Col2a1+Prg4+) and synovial and ligament fibroblasts (Prg4+ and/or Scx+).
Translational Research
Degenerative joint disease, also known as osteoarthritis, is among the top 5 most costly medical conditions in the Unites States. One of the challenges of repairing the articular cartilage that lines our joints is that this tissue forms prenatally, and regeneration does not normally occur after birth. We hypothesize that a better understanding of how joint tissues arise during embryonic development will enable us to develop better approaches for treating damaged joint tissues in patients. To this end, we developed methods to generate articular cartilage tissues from human pluripotent stem cells, and showed that they effectively repaired cartilage defects in a small animal model. We are progressing now towards preclinical large animal studies of cartilage repair, as a prelude to repairing damaged tissue in patients.
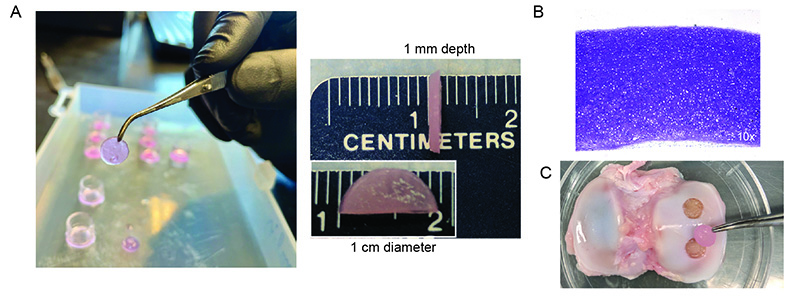
Following successful repair of rodent joint cartilage (Gardner et. al, 2019), we are excited to continue our translational studies in a large animal model. (A) We are engineering cartilage with defined thickness and diameter for preclinical studies. (B) Histological cross sections of articular cartilage generated from hESCs stains metachromatically with toluidine blue dye indicating the presence of proteoglycans. (C) Example of minipig joint surface (tibial plateau) in which we created full thickness focal cartilage defects (5 mm diameter). Held in forceps is a size-matched biopsy punch of a hESC-derived cartilage tissue construct created in the lab.